SciLake at FENS Regional Meeting 2025
SciLake at FENS Regional Meeting 2025
The Federation of European Neuroscience Societies (FENS) Regional Meeting 2025, held from June 16-19 in Oslo, Norway, brought together researchers, clinicians, and students from across the neuroscience community. The conference covered a broad spectrum of topics, from fundamental research to clinical applications, with special focus on the integration of artificial intelligence and computational models in neuroscience research.
SciLake Representation
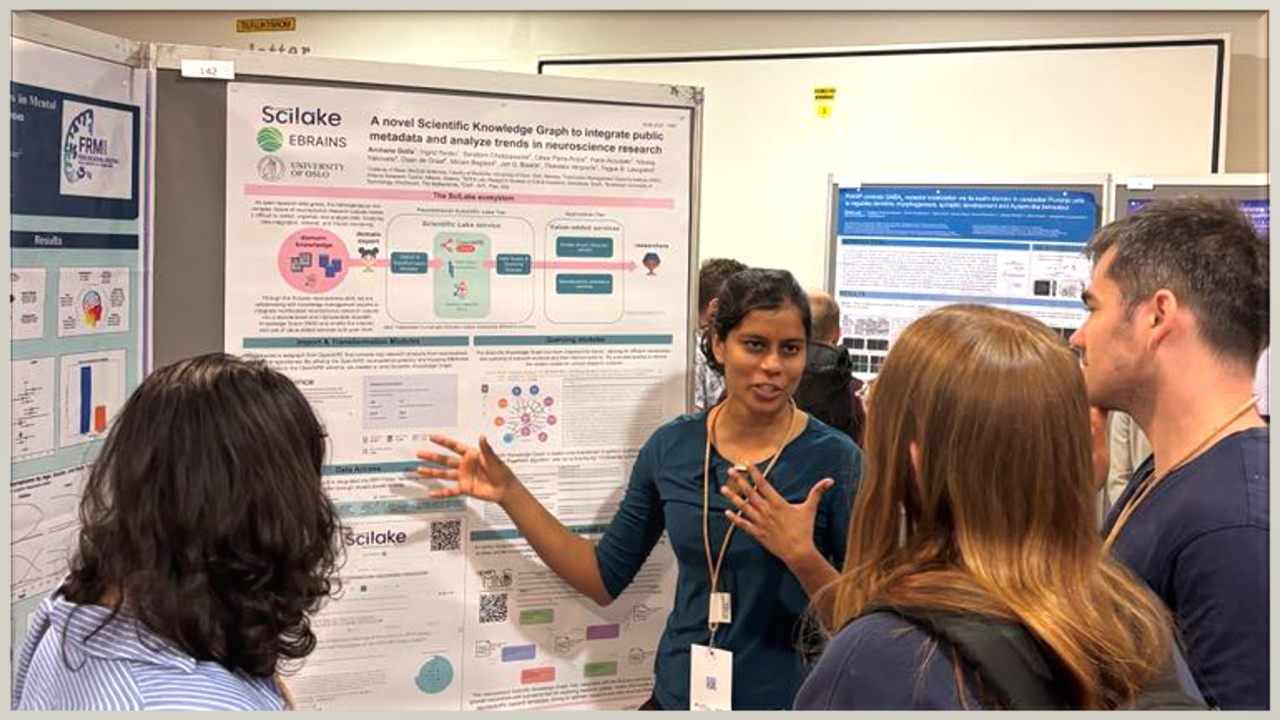
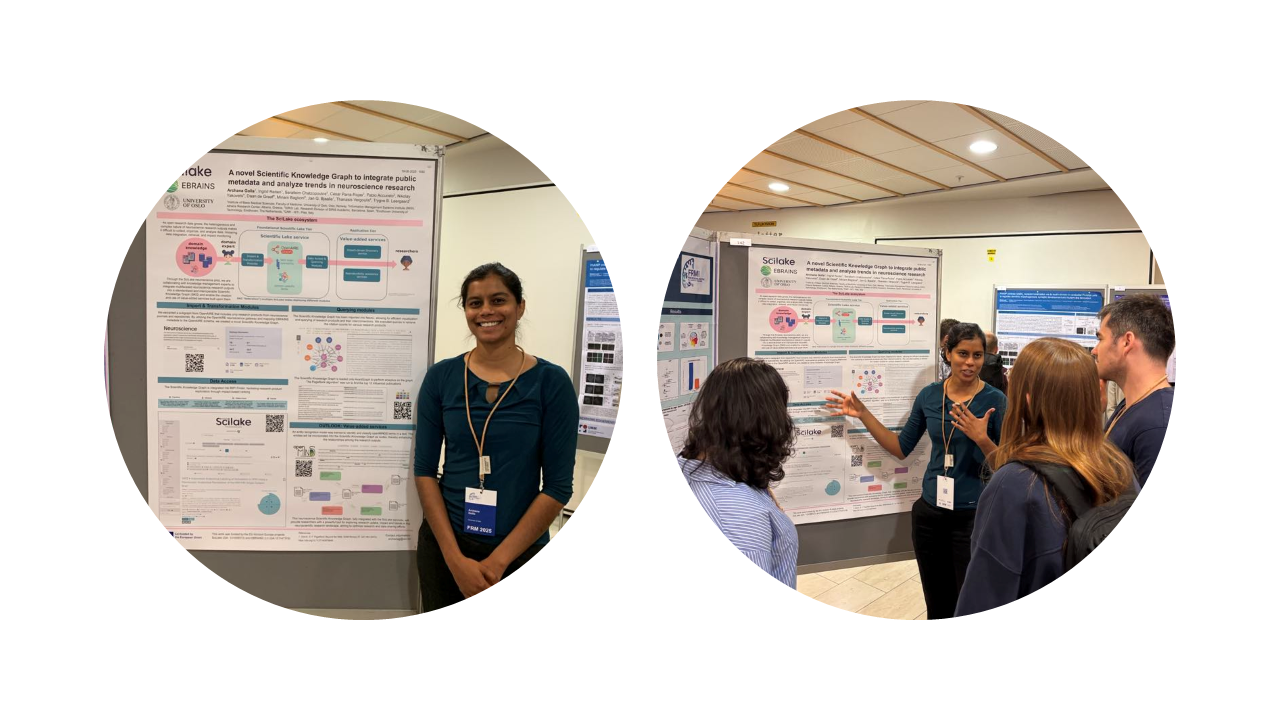
Archana Golla from the University of Oslo represented the SciLake project, presenting a poster on our innovative Scientific Knowledge Graph (SKG) developed specifically for neuroscience research.
The poster titled "A Novel SKG to Integrate Public Metadata and Analyse Trends in Neuroscience Research" showcased SciLake's work in addressing the challenges of organizing and analyzing the growing volume of heterogeneous neuroscience research outputs.
Technical Innovation
Our neuroscience pilot has established a novel SKG by integrating:
- Scholarly metadata automatically harvested from journal publications in the OpenAIRE Graph
- openMINDS metadata from EBRAINS datasets mapped to the OpenAIRE metadata schema
Key Features of the SciLake Neuroscience SKG
The SKG offers several advanced capabilities:
- Implementation in AvantGraph to facilitate advanced citation analyses
- Integration of SIRIS entity recognition tools to establish connections between datasets and publications
- Programmatic access and user-friendly interface highlighting linked datasets and publications
- Comprehensive citation metrics to track research impact
Impact and Applications
The neuroscience SKG provides researchers and service providers with a powerful tool for:
- Exploring research uptake and impact
- Identifying emerging trends in neuroscience research
- Optimizing research and data sharing efforts
- Improving the visibility and accessibility of valuable datasets
Stay in touch
Following the positive reception at FENS 2025, the SciLake team will continue refining the SKG and expanding its capabilities. We invite researchers interested in exploring or contributing to this innovative tool to contact us through the project website: https://scilake.eu/neuroscience-case-study