Amplifying Valuable Research: How?
Amplifying Valuable Research: How?
In today's fast-paced scientific landscape, it has become increasingly challenging for researchers to identify valuable articles and conduct meaningful literature reviews. The exponential growth of scientific output, coupled with the pressure to publish, has made the process of discovering impactful research a daunting task.
In a webinar for SciLake partners,Thanasis Vergoulis, Development and Operation Director of OpenAIRE AMKE and scientific associate at IMSI, Athena Research Center, discussed the technologies developed to assist knowledge discovery by leveraging impact indicators as part of SciLake's WP3:Smart Impact-driven Discovery service.
This article recaps the key points from the webinar.
Impact Indicators and knowledge discovery
Scientists heavily rely on existing literature to build their expertise. The first step is to identify valuable articles before reading them. Unfortunately, this process has become increasingly tedious due to the overwhelming volume of scientific output. The increasing number of researchers and the notorious publish-or-perish culture have contributed to this exponential growth, making it difficult and time-consuming to identify truly valuable research in specific areas of interest.
Impact indicators have been widely used to address this challenge. The main idea is to look at how many articles cite a particular article, which serves as an indication of its scientific impact. This is formalized through thecitation countindicator.
Thanks to the adoption of Open Science principles, there is now a wealth of citation data available from initiatives like Open Citations and Crossref. As a result, the now-available citation data offer adequate coverage to estimate the scientific impact of an article analyzing its citations. Of course, scientific impact is not always highly correlated with scientific merit, hence it is always important to remember that an article of great value might not always be popular.
While citation count is a popular impact indicator used in academic search engines like Google Scholar, it has its limitations. Scientific impact is multifaceted, and one indicator alone is not sufficient to measure it. Other pitfalls related to citation count, that may hinder the discovery of valuable research, are the introduction of biases against recent articles and the potential for gaming the system by attacking this indicator with particular malpractices. To mitigate such issues, it is crucial to use indicators that capture a wide range of impact aspects. Additionally, considering indicator semantics and provenance helps protect against improper use and misconceptions.
How it started…
To study this problem, researchers fromAthena Research Center (ARC) conducted acomprehensive survey and a series of extensive experiments to explore different ways to calculate impact indicators and rank papers based on them. Four major aspects of scientific impact were identified that should be combined:
- Traditional impact, estimated with citation counts
- Influence, estimated using thePageRank algorithm, which considers the impact of an article even if it is not directly cited
- Popularity, estimated using theAttRank algorithm, which considers the fact that recent papers have not had sufficient time to accumulate citations
- Impulse, estimated using 3-year citation counts to capture how quickly a paper receives attention after its publication
Building upon these aspects, ARC researchers developed a workflow to calculate these indicators for a vast number of research products and made them openly available to the community, enabling third-party services to be built on top of them.
…how it is going
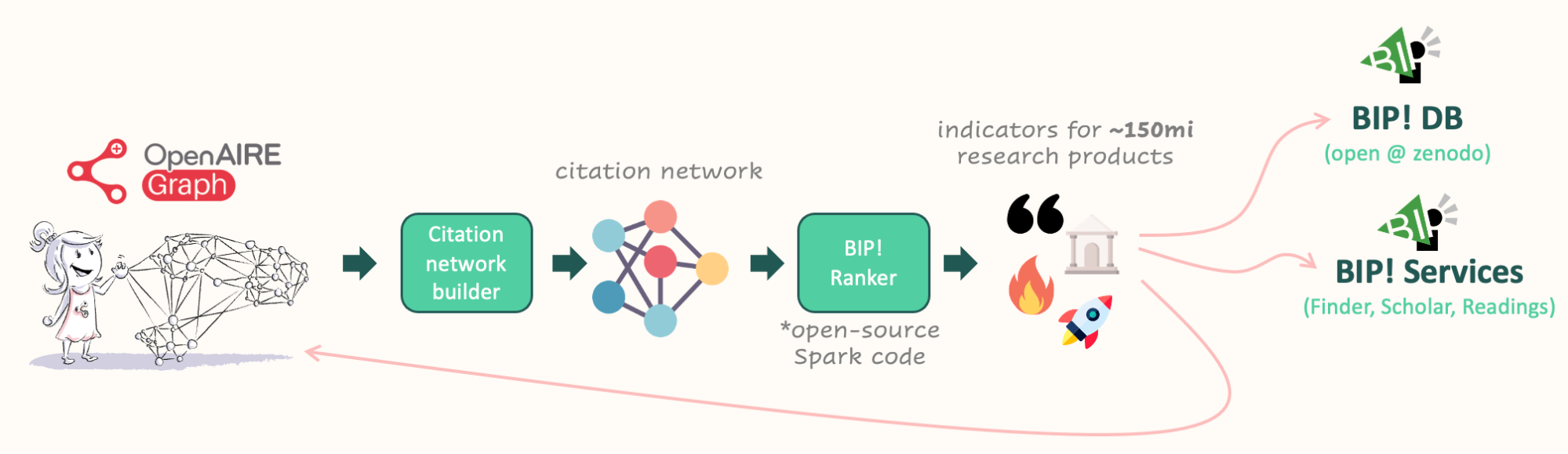
The workflow to the BIP! Database and Services
The current workflow developed by ARC starts with the OpenAIRE Graph, where citations are collected as a first proxy of impact. A citation network is built based on this information. ARC has developed an open-source Spark-based library called BIP! Ranker, which calculates indicators for approximately 150 million research products. While computationally intensive, the calculations can be performed within minutes or hours on a computer cluster, depending on the indicator. The resulting indicators are available on Zenodo as the BIP! DB dataset and advanced services, such as BIP! Finder, BIP! Scholar, and BIP! Readings are also provided based on these indicators. Finally, the indicators are integrated back into the OpenAIRE Graph, ensuring their inclusion in any data snapshot of the graph downloaded. In addition, the workflow classifies research products based on their ranking and can provide, for example, the results within certain percentage thresholds, (e.g. being in the top 1% of the whole dataset or of a particular topic). During the process of calculating the indicators, various checks should take place. For instance, to prevent the duplication of citations, it is ensured that multiple versions of the same article, such as pre-prints and published versions, are not counted twice. Additionally, there are plans to eliminate self-citations in the future and to give the option to select whether to consider only citations from peer-reviewed articles or not.
So, what can we use?
BIP! Finder: the service that improves literature search through impact-based ranking

In the BIP! Finder interface, users can perform keyword-based searches and rank results based on different aspects of impact (e.g., popularity or influence). This allows users to customize the order of the results. Each result also displays the class that each publication has according to the four main impact indicators available through the interface. The service also provides insight into how a paper is ranked among others in a specific topic. This is particularly useful in cases of highly specialized papers which would unlikely rank high in a large database.
Preparations for SciLake pilots
The BIP! services bundle now includes theBIP! spaces service that allows building domain-specific, tailored BIP! Finder replicas. The main purpose of these spaces is to use them as demonstrations for the pilots of the SciLake project. The service will provide knowledge discovery functionalities based on impact indicators and incorporating information from the domain-specific knowledge graphs that the pilots are building.
What each pilot gets:
- a preset in the search conditions, such as the preferred way to rank the results,
- query expansions with additional keywords based on domain-specific synonyms (e.g., synonyms in gene names in cancer research),
- query results including domain-specific annotations based on pilots' scientific knowledge graphs.
Future extensions:
- support for annotating publications to extend the domain-specific SKGs:
- enabling users to add connections of publications and other objects to domain-specific entities and include these relations into their SKG,
- additional indicators,
- support for domain-specific highlights:
- flags for collections of papers that are important in a specific community,
- topic summarization & evolution visualisation features.
Conclusions
By leveraging impact indicators, researchers can navigate the vast scientific landscape more effectively, discover valuable research, and make informed decisions in their respective fields. This paves the way for accelerating knowledge discovery and amplifying the impact of valuable research.
Stay tuned for more updates on how SciLake is amplifying valuable research!