SciLake at ISMB/ECCB 2025
Advancing Cancer Knowledge Graphs for Precision Oncology
By Leily Rabbani, Richard Rosenquist, Stefania Amodeo
The International Conference on Intelligent Systems for Molecular Biology (ISMB), jointly held with the European Conference on Computational Biology (ECCB) 2025, took place in Liverpool, England from July 20-24, 2025. The event brought together approximately 2,000 participants from the global bioinformatics and computational biology community, including researchers, students, industry experts, and academics.
SciLake was proudly represented by Leily Rabbani from Karolinska Institutet, who attended on behalf of the Cancer pilot (WP5). This blog post summarizes our participation and the presentation of our SciLake’s Cancer Knowledge Graph.

Conference Highlights
ISMB/ECCB 2025, organized by the International Society for Computational Biology (ISCB), featured a rich program of keynote talks, specialized sessions, workshops, and poster presentations that showcased advances in computational biology.
A significant focus this year was on artificial intelligence (AI) and large language models (LLMs) in computational biology and molecular research. These themes are particularly relevant to Scilake’s interests, highlighting innovative applications of AI-driven approaches in genomics, structural biology, and data analysis.
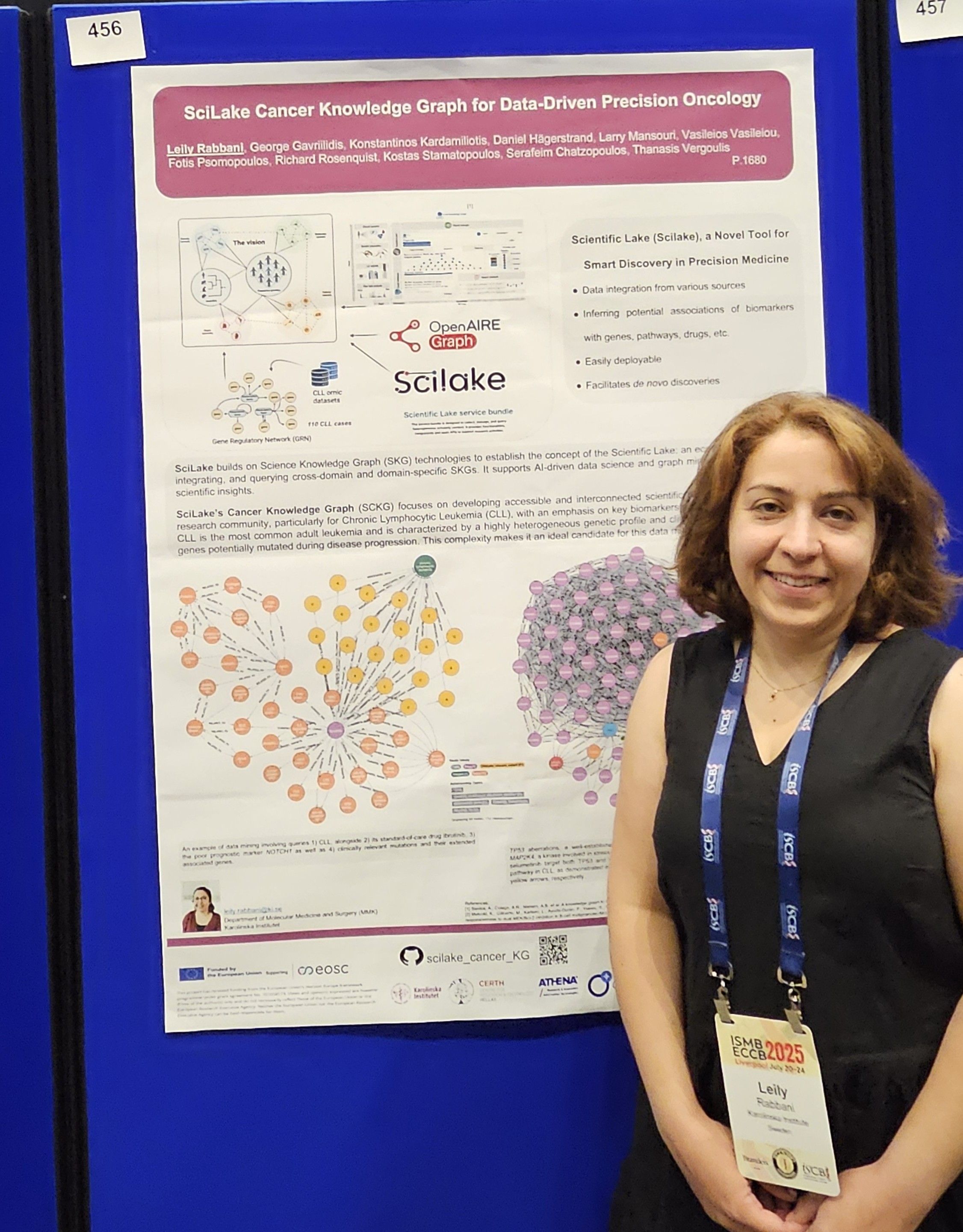
SciLake's Cancer Knowledge Graph
Our team presented a poster titled "SciLake Cancer Knowledge Graph for data-driven precision Oncology," authored by Leily Rabbani (presenter), George Gavriilidis, Konstantinos Kardamiliotis, Daniel Hägerstrand, Larry Mansouri, Vasileios Vasileiou, Fotis Psomopoulos, Richard Rosenquist, Kostas Stamatopoulos, Serafeim Chatzopoulos, and Thanasis Vergoulis.
The research attracted considerable attention from conference attendees, particularly for its innovative approach to developing accessible and interconnected scientific resources for cancer research. Our knowledge graph specifically targets the identification of key biomarkers and potential druggable targets, addressing critical needs in precision oncology.
Technical Framework
Built upon established frameworks including the Clinical Knowledge Graph, OpenAIREGraph, and Gene Regulatory Networks from cancer-specific multiomics datasets, the SciLake's Cancer Knowledge Graph creates comprehensive connections between genes, proteins, metabolites, drugs, and scientific publications.
Discoveries that Matter
Our pilot case explores chronic lymphocytic leukemia (CLL), the most common adult leukemia. We built our knowledge graph using data from the BloodCancerMultiOmics2017 dataset, analyzing patterns in 111 patient cases (55 aggressive and 56 indolent).
The resulting CLL knowledge graph contains over 14 million nodes and nearly 200 million relationships.
To demonstrate the practical value of our tool, we investigated TP53, a gene known to predict poor outcomes in CLL patients. Our knowledge graph revealed a previously underexplored relationship between TP53 and MAP2K4, a gene involved in cellular stress response.
This analysis identified three existing medications (mirdametinib, trametinib, and selumetinib) that target both genes, providing evidence that the MAPK/ERK pathway plays a significant role in CLL development - a finding that aligns with previous laboratory studies.
What's Next for Our Research
We are making our cancer knowledge graph even more powerful by adding:
- AvantGraph SKG-engine - to enhance how researchers can explore the data
- BIP! Spaces - to automatically extract information from scientific papers
- Gene Regulatory Networks built from different types of cancer data (including single-cell RNA sequencing and protein analysis).
Our participation at ISMB/ECCB 2025 provided valuable opportunities for networking, collaboration, and exchange of novel computational methods and biological insights, supporting both established scientists and early-career researchers.