Unlocking insights in Cancer Research through Knowledge Graphs
In a recent meeting with the EOSC4Cancer Cancer Landscape Partnering (CLP), SciLake took center stage as it introduced its vision and roadmap for unlocking insights in cancer research. Project coordinator, Thanasis Vergoulis (Athena RC), and Leily Rabbani, bioinformatician at the Department of Molecular Medicine and Surgery in Karolinska Institute, discussed the ongoing work towards creating a Cancer Research Knowledge Graph. This innovative tool will provide context and connections for what is known about specific research questions, helping researchers as they design new experiments.

The SciLake Cancer Research pilot involves the Institute of Applied Bioscience (INAB-CERTH) in Greece and Karolinska Institutet in Sweden. Focused on meeting the needs of researchers and clinicians, the project aims to harness the wealth of information available in public resources to address ongoing research questions.
The ultimate goal? To deepen our understanding of the molecular biology and immunopathology of Chronic Lymphocytic Leukemia (CLL) and study the potential effects of different mutations.
With the assistance of SciLake technical partners, members of the pilot project are utilising advanced algorithms to discover new insights from the knowledge graph. For example, one interesting question they are exploring is, "how might a specific genetic mutation forecast a patient's overall health status and what insights might related literature offer in this regard?"
Chronic Lymphocytic Leukemia (CLL)
- Characterized by the accumulation of neoplastic B cells in the bone marrow, blood cells, and secondary lymph nodes.
- Patients can have a very diverse genetic landscape leading to heterogeneous clinical outcomes. This means progression rates and responses to drugs can vary greatly among patients.
- The most common type of leukemia in adults.
- Currently incurable.
Knowledge Graph: Benefits
The use of a scientific knowledge graph offers several benefits. It empowers research in precision medicine and diagnostics by facilitating the discovery of potential associations between identified biomarkers and other elements, such as genes, biological or functional pathways, and drugs. Furthermore, it is easily deployable and flexible, capable of integrating data from various sources, thereby offering a comprehensive view of the research landscape.
Challenges
Developing a knowledge graph comes with its own set of challenges. The objective is to provide tools for creating and enriching the graph, and the primary concern is extracting latent knowledge to create the graph. Another significant challenge is establishing a common language among people of different expertise, such as clinicians and technical developers. This is crucial to facilitate effective communication and collaboration in the development and application of the knowledge graph. Finally, an important step to validate the graph involves manual curation to assess hidden associations and existing connections and ensure they are relevant to the specific biology experiment.
Where are we
The development of the knowledge graph is progressing by leveraging several pre-existing state-of-the-art knowledge graphs. One is PrimeKG, which is used to query networks of genes or proteins connected to a specific disease. For example, the graph shows connections between CLL and TP53, a gene known for its potential to increase the risk of various forms of cancer significantly when altered. Other larger state-of-the-art knowledge graphs, based on various biomedical databases, are also being integrated along with Prime KG. This strategy aims to capitalise on a broader set of databases and underlying connections, potentially uncovering new missing links. An example of this is the revealed relation between CLL and the gene SOD1, known for being overexpressed in many human cancers.
A variety of knowledge graphs exist, each drawing from a different biomedical source, and we can collect more information through their combination. In fact, many details are unique to a particular graph and there is minimal overlap between them.
Our data flow involves using a variety of knowledge graphs, including those previously mentioned, along with different ontologies and other data sources. We utilize tools provided by SciLake to establish connections among them and generate a comprehensive cancer knowledge graph.
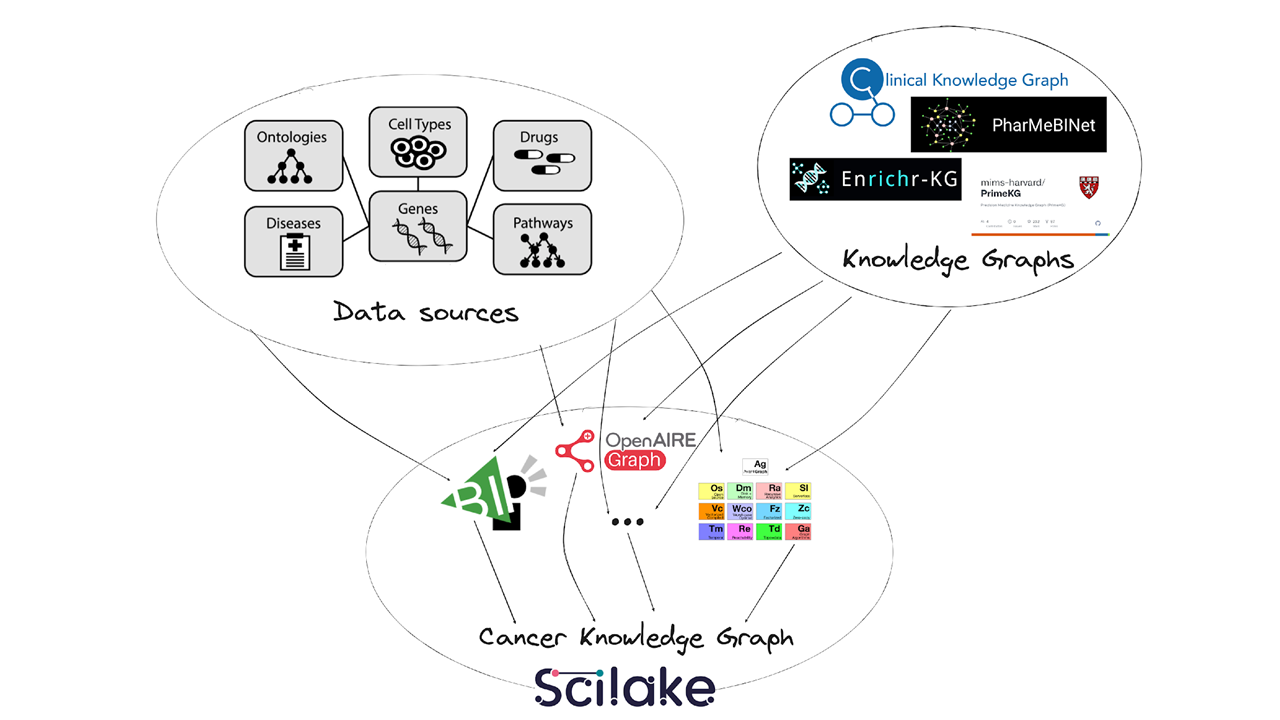
Dataflow towards a CLL KG
Goals
The ultimate vision is to create a comprehensive network of interconnected nodes and relationships. These nodes can represent various entities such as institutions, grants, patents, publications, software, anatomical structures, diseases, drugs, compounds, gene targets, and many more. These relationships can take on different forms and can signify different types of connections such as mentions, associations, or other types of relationships. By creating this extensive web of connections, the network can be navigated and queried in a semi-automated manner to answer specific research questions. Moreover, the impact and reproducibility analysis services offered by SciLake can be utilized to prioritize findings.